GCCA_tsdata_to_mvgc
Calculate conditional time-domain MVGC (multivariate Granger causality) from time series data by "traditional" method (as e.g. in GCCA toolbox)
Syntax
F = GCCA_tsdata_to_mvgc(U,x,y,p,regmode)
Arguments
See also Common variable names and data structures.
input
U multi-trial time series data x vector of indices of target (causee) multi-variable y vector of indices of source (causal) multi-variable p model order (number of lags) regmode regression mode (default as in 'tsdata_to_var')
output
F Granger causality
Description
Returns the time-domain MVGC
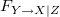
from the variable Y (specified by the vector of indices y) to the variable X (specified by the vector of indices x), conditional on all other variables Z in the time series data U. The regression mode is set by the regmode parameter, which may be 'LWR' or 'OLS' (see tsdata_to_var for details and defaults).
If p is a vector of length 2, then p(1) is the number of lags for the full regression and p(2) the number of lags for the reduced regression. Otherwise, if p is a scalar, p lags are used for both the full and reduced regressions.
This routine is included mainly for compatibility with the Granger Causal Connectivity Analysis (GCCA) Toolbox [2]; the MVGC is calculated by performing separate full and reduced regressions - see [1] for details. Note that the preferred MVGC Toolbox method of calculating MVGC via the autocovariance sequence (see autocov_to_mvgc, mvgc_demo) only requires a single regression step and is thus generally more accurate.
The caller should take note of any warnings issued by this function and test results with a call isbad|(F,false)|.
References
[1] L. Barnett and A. K. Seth, The MVGC Multivariate Granger Causality Toolbox: A New Approach to Granger-causal Inference, J. Neurosci. Methods 223, 2014 [ preprint ].
[2] A. K. Seth, "A MATLAB toolbox for Granger causal connectivity analysis", Journal of Neuroscience Methods 186, 2010.