Discrete Dynamics Lab
Update Feb 1999 -
version 22
- new features
New features added since the last DDLab release
in Sept 1997 are listed below, with links to illustrations.
The list is not exhaustive.
A number of logical, presentaton and "crash"
bugs have also been fixed (and not too may added - I hope).
Previous updates, Sept
1997 since March 96, are listed
here.
An updated manual covering all DDLab functions,
with examples and illustrations, is in the process of
being written.
New features and changes
3d networks
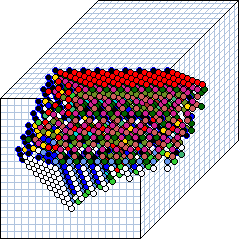
- 3d networks are now implimented (see examples
1
and
2).
- Most functions for
1d and 2d networks also apply to 3d,
for both cellular automata (CA)
and random Boolian networks (RBN).
- The maximum size
of network supported is 64k, giving a maximum cube of
40x40x40, or equivalent non-cubic space.
- To impliment 3d networks, various new features are introduced,
including defining 3d local neighbourhoods, a new 3d wiring
graphic, methods for setting and revising the wiring and the
initial state in 3d, displaying space-time
patterns in 3d, and saving the 3d network.
The local neighbourhood
- The local neighbourhood for CA, and pseudo-neighbourhood
for RBN, is defined in
3d,
in a similar way to
2d
and
1d.
- The neighbourhood for 2d and 3d has been defined to be
as symmetric as possible, so that for
even k there is no self wiring.
- In 2d networks, both k = 6 and 7 define an effectivly
triangular grid.
The wiring graphic
- The 3d wiring graphic shows cells as connected colored spheres,
as if looking up into an axonometric "cage"
(
see examples for 1d, 2d and 3d),
allowing the wiring (and rule) to be displayed
for specified network cells, with a range of further options
to ammend the wiring (and rule) for that cell, or for a specified
subset of cells.
The methods are analagous to those for 2d and 1d.
- For 3d networks, both a 2d and 3d wiring graphic are shown
simultaneously
( see example).
The 2d graphic divides the 3d space into labeled horizontal
slices (layers) usualy resulting in tall rectangle.
- The 2d wiring graphic can be moved up and down
to scan the entire network if part is off the screen;
important for large 3d networks shown in 2d.
- The 2d and 3d wiring graphics can be independently
expanded/contracted.
- The wiring graphic can be displayed in
1d.
irrespective of the "native" dimention of the network.
- Presentation (for 1d, 2d and 3d) can be toggled:
with/without pseudo nhood, with/without links
(see 3d example).
New flexible biases for "random" wiring
- A subset of cells as well as the
current cell can be designated in the wiring graphic.
(3d-layers 2d-rows 1d-range), so that
wiring (and rule) biases will apply to all the cells
in the subset rather than just one cell.
- When random wiring is confined to a local zone,
up to k wires
can be released from the zone.
- The default periodic boundary conditions can be seperatly
suppressed on the relevant axes for 3d, 2d and 1d networks, i.e.
the i,j,h axes for 3d networks.
- Links to particular layers in a 3d network,
rows in 2d, or a range in 1d, can be disallowed.
- "Direct" links can be forced to a particulr layer in a 3d network.
- A given number of random links can be forced to different
layers in a 3d network.
- For uniform k, the same random wiring template
can be appled to all network cells (quasi CA wiring).
Untangling wiring
- A cell's wiring can be
"
untangled"
so that wires do
not cross relative to the pseudo-neighbourhood as
shown in the 1d wiring graphic. This applies
to networks of any "native" dimention (1d, 2d or 3d).
- Untangling can be done to a single cell, or to the whole
network.
- Untangling is a neutral transformation in a mixed rule
network. If the mixed rules have been set, a cell's rule
is transformed so that the change in wiring has no effect
on the dymamics. In a single rule network, the untangling
doed not change the rule so is not neutral.
Mixed k and effective k
- After setting the k-mix, a new option allows the
max-k to be set greater than the maximum k
in the network. This allows individual k's to
be subsequently increased up to max-k.
- A new algorithm allows "effective" k to be found
for particular cells or for the whole network (assuming
a mixed-k network). A given rule may cause some
inputs to be "neutral", having no effect on the dynamics.
The neutral imputs can be removed, reducing the cell's
wiring to the minimum required, and changing the rule
accordingly.
- For a k-mix network, k can
be neutraly ncreaced (up to max-k), and effective
k can be found,
for a single cell, or for the whole network.
The inverse problem
- An "inverse problem" method first finds the the fully
connected network (k=n)
corresponding to an "exaustive map"
(basin of attraction field).
The k=n network then has its connections reduced
to "effective k". The algorithm may be tested by
generating the exhaustive map from known simple
RBN or CA. The idea for this method is due to John Myers.
Indirect inputs and outputs - degrees of separation
- From the 1d wiring graphic, a new option displays all
inputs and outputs to a given cell, direct and indirect.
These are shown all at once or in steps (degrees of separation).
An
example is given for an RBN,
k=2, and 6 degrees of separation.
The number of cells effected is given, and
shown in a 2d representation of the network.
Setting rules
- For mixed rule networks, a subset of cells
can be designated in the wiring graphic
(3d-layers 2d-rows 1d-range).
A specified rule can be copied to all the cells
in the subset.
- A number of new options have been added
to "transforming rule/s", which may apply to
all the rules in a mixed rule network, or just to one rule.
- "Solidify": so that "on" cells (i.e. with value 1) stay on.
- "Invert": transform rule/s between the
two common conventions for arranging the rule-table:
in descending order of the neighbourhood value,
from left to right (the convention in DDLab),
or from right to left, a common alternative convention.
- Set "canalizing".
- As well as tranforming rule/s to greater k,
rule/s (and wiring) can be transformed to "effective"
k by pruning unnecessary inputs.
- There are a number of changes and new options
for setting single rules:
- The rule table can be set to all 1s, as well as all 0s,
for subsequent editing of bits.
- A rule bias for CA suggested by Lee Altenberg (designated "Alt"),
where the output of each
neighbourhood in the rule table is set to 1 with a
probability depending an the density of 1s in the neighbourhood.
This often results in co-existing domains, gliders etc.
- The Life rule is now set with "life-L" (capital L).
- The rule-table can be biased to have a preset density of 1s
(i.e. lambda parameter).
Setting the seed (initial state)
- As in the "wiring graphic", in setting the initial state for
3d networks, both a 2d and 3d seed graphic can be shown
simultaneously (when setting bits with the mouse or
keyboard). The 3d seed graphic shows "on" cells as spheres,
as if looking up into an axonometric "cage".
The 2d seed divides the 3d space into labeled horizontal
slices (layers) usualy resulting in a tall rectangle.
(
see example).
- "Bits" can be "drawn" with the mouse or keyboard on the
2d seed graphic in the usual way; the 3d graphic can be
immediately updated.
- The 2d graphic can be moved up and down
to scan the entire network if part is off the screen;
important for large 3d networks shown in 2d.
- The 2d and 3d seed graphics can be independently
expanded/contracted.
- The 3d seed pattern can be "rotated",
i.e. moved along the 3 periodic axes.
- When setting bits in 2d, vertical (as well as
horizontal) lines can be drawn from the keyboard.
- The pattern can be complimented.
- The density of 1s in a random seed, across the whole network,
or in a central block (1d, 2d or 3d) can be biased. The density
can be "strict" or according to a probability.
Space-time patterns
- The 3d space-time pattern generaly shows "on" cells as
spheres, as if looking up into an axonometric "cage",
unless the scale is very small
(
see example).
- New "
on the fly" options include the following:
- Space-time patterns can be toggled between a 1d-2d-3d display.
Given a "native" dimension other than 3d, the 3d
coordinates are set automatically.
- A 3d network in 2d shows horizontal slices (layers)
stacked above each other
(
see example).
- 2d space-time patterns can be toggled between
2dand
2d+time as before,
whatever the "native" dimension.
-
Filtering CA, see below.
- The
frequency
of "on" cells (having value "1") within a
moving time window can be displayed according to a color code.
- Toggling the cell color according to different criteria
has been refined. As well as showing cells colored
according to
value or neighbourhood lookup,
the display can be toggled between the following
(with color key),
where "frozen" indicates cells that have not changed value
for a predetermined number of time-steps
(
see example) .
1. normal i.e. 1s-black 0s-white (or equivalent).
2. frozen cells-red, rest norma3.
3. frozen 0s-green, frozen 1s-red, rest white.
4. color coded according to the frequency of 1s.
- "Time" can be compressed by not showing alternate
time-steps, a preset number of time-steps, between
displayed iterations.
- The space-time pattern need not be shown to speed up
other data gathering.
- A pause can be set at each time-step.
- The itteration of the space-time pattern can be slowed down.
- 'Q' interrupts without displaying the large option window
(and hiding graphics).
- A pause can be set after N preset time-steps.
The pause can be continued if required.
- The "entropy" dispay can toggle between
just the entropy,
just the lookup-frequency traces,
or both.
- The current state, or original seed, can be saved.
Filtering CA space-time patterns
CA can be
"
filtered"
to shows up gliders or domain
boundaries. This works by not displaying network elements
that updated with refecence to the most
frequently "looked up" neighbourhoods in a moving window of
time-steps.
- Neighborhoods are filtered sequentialy from the
most frequent down. Alternativly, selected
neighourhoods can be filtered.
- A dot is displayed alongside the filtered
neighbourhood in the lookup frequency histogram
(
see example.
- Unfiltering is done sequentially in reverse order,
or all at once.
- Filtering works in all modes, by neighbourhood color,
by value, or the various
"frozen" modes, or frequency of 1s color code.
- However, the lookup-freq hist must be on (default for 1d),
i.e. must be set for and 2d and 3d.
Attractor Basins
- States belonging to basins of attraction can be saved,
in addition to other attractor basin data.
In the the new data format, a typical item in the list
"101011 2 0 2" signifies the following:
101011 - the state shown as a bitstring.
2 - basin number.
0 - 0 or 1, 0 = transient state, 1 = attractor state.
2 - the number of pre-images of the state.
i.e. 0 = a garden-of-Eden state.
- A count can be made of
states with a density of 1s greater or less than .5 in
subtrees, basins of attraction or the entire field.
This can be applied to check the
fittness of rules for the "density problem" in
"emergent computation".
- A graph of G-density against net size n for
increasing n can be generated, for subtrees,
basins of attraction or the entire field..
Derrida plots
- A number of refinements have been made, including the
following
(
see example):
- Parameters set become new defaults.
- The number of itterations is shown on the plot.
- Canalizing can be changed between successive Derrida plots.
- Previous graphs can be kept to show multiple overlays.
Input entropy rule samples
- Samples of rules sorted according to
entropy standard deviation - mean entropy are included
in the release for k=5,6 and 7.
- The
scatter plots
are now shown according to a color code
indicating log2 of frequency. i.e. colors change for
1,2,4,8 etc., corresponding to the plot shown as a
frequency histogram.
- A point on the scatter plot can now be probed for lambda
and Z as well as mean entropy.
- The rules at each point can be listed, and a rule selected
to see space-time patterns.
Presentation and Operation Changes
- New option to change font size.
- Inputs to all prompts can be revised with "q" as well as
"Backspace"
- DOS:
- Screen resolution of 800x600 can be selected
(as well as well as 640x480 and 1024x768).
- Only the 640x480 runs in a Windows95 window
(alt-return toggles between the full screen and a window).
- Higher resolution takes up the full
screen. Running from DOS rather than Windows is
recomended.
Return to the
Discrete Dynamics Lab home page.